Deep dive: Plant-based meat crop development
Learn about the ingredients in plant-based meat, the crops they come from, and how crop innovations can benefit the development of plant-based products.
Crop development
Introduction
Plant proteins serve as the primary raw material for plant-based meat. Optimizing their crop sources results in less costly, energy-demanding, time-consuming, and complex downstream processing. The more closely tailored a raw plant protein is for plant-based meat, the less effort is needed to achieve the desired functional and sensory aspects through subsequent steps in the process, such as flavoring, formulation, and mechanical production.
Historically, most crops used as the predominant protein sources for plant-based meat have been optimized for use of their oils and starches, but crop varieties geared toward protein products must be developed. Common plant-protein sources, such as soy and wheat, are used simply because they exist in abundance as side streams of other processes. As GFI’s plant protein primer and GFI APAC’s Asian Cropportunities report emphasize, many other plant-protein sources remain underutilized and underexplored (see Table 1).
The primary starting material for plant-based meat production is typically plant protein concentrate (comprising 60%–70% protein) or plant protein isolate (composed of at least 90% protein). These proteins form the major ingredient of the end product, with smaller fractions comprising other components, such as lipids, starches, and flavoring agents. This deep dive focuses on the structures and functionalities of relevant plant proteins and identifies prospects for improving their sources. The ingredient-optimization and end product formulation and manufacturing deep dives discuss opportunities to improve isolation and processing of these proteins for plant-based meat applications.
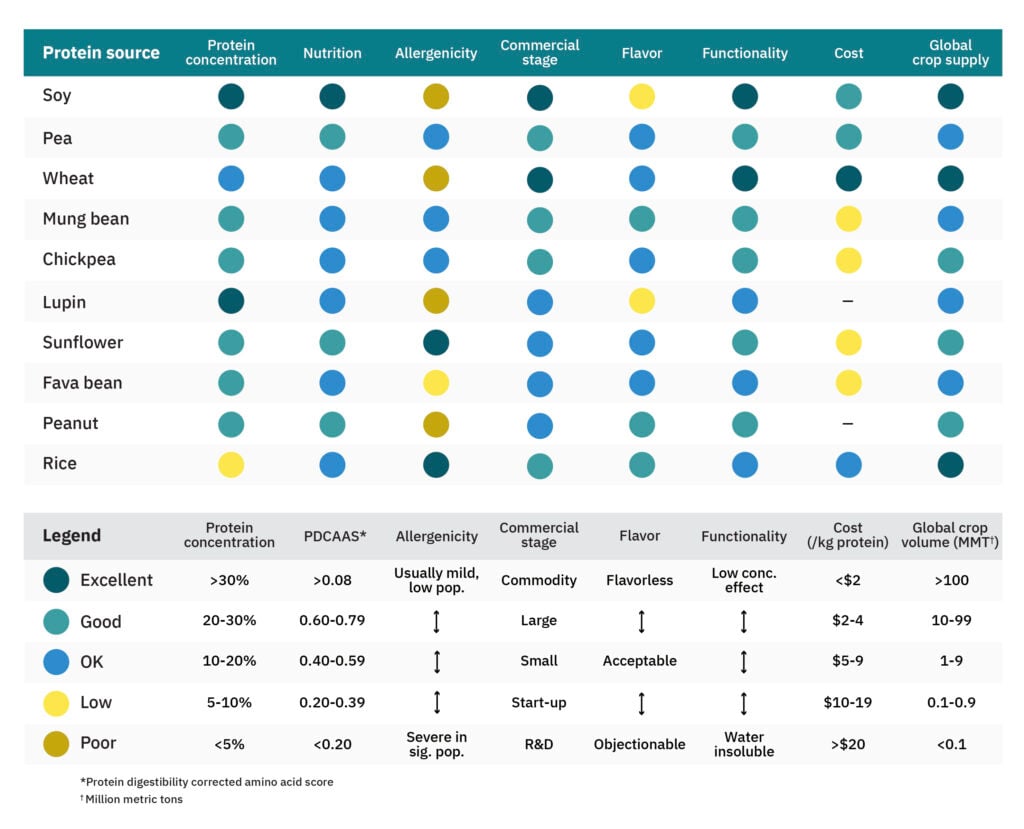
Note: Of the top 10 U.S. plant-based meat retail brands, nine use soy, wheat, or pea protein despite the compelling properties demonstrated by other proteins.
Identifying and characterizing plant proteins with promising functionalities
Creating plant-based meat entails crafting products that mimic or improve the taste, texture, nutrition, and overall experience of animal-based meat. Accordingly, understanding the differences between plant and animal protein characteristics and functionalities is crucial. The distinctions stem from the amino acids (AAs) that make up plant and animal proteins, their sequences, and their interactions with one another to form protein structures. Ultimately, proteins’ compositions, sequences, and structures affect their solubility, thermal stability, isoelectric points, nutrition profiles, texture, water- and fat-holding capacities, emulsifying and foaming capabilities, and gelling abilities. This section aims to explore the sequences and structures of plant and animal proteins. The functionality of proteins and nonprotein plant-based raw ingredients is also discussed here. Please refer to GFI’s Formulating with animal-free ingredients and end product formulation and manufacturing deep dive for more in-depth considerations of plant protein formulations.
Protein amino acid composition and digestibility
AAs are the basic building blocks of proteins. Optimal AA concentration and balance are desired to improve meat products’ nutrition, texture, and taste. Human bodies can synthesize 12 of the 21 AAs contained in proteins. The nine AAs that we cannot make must come from our diets and are dubbed essential amino acids. On this basis, World Health Organization et al. 2007 and the U.S. Institute of Medicine 2005 developed recommendations for daily AA intake. Animal proteins offer balanced and highly concentrated essential AA compositions. Plants can synthesize all AAs necessary to make proteins, but native crop proteins often lack adequate essential AA scores and are less digestible than animal proteins (Gorissen et al. 2018). For instance, wheat gluten and rice are low in lysine, while legumes tend to be low in the sulfur-containing AAs methionine and cysteine. Moreover, plant protein structures make the proteins more resistant to proteolysis in the gastrointestinal tract. Fibers, antinutritional factors, and other impurities in plant protein products can also lower protein digestibility. Note that, although plant proteins tend to be less digestible than animal proteins, a recent study revealed that replacing animal meat with plant-based meat promotes positive changes to gut microbiomes (Toribio-Mateas et al. 2021).
Quantitative measurements of protein digestibility help compare protein quality. Commonly applied methods include the protein digestibility corrected amino acid score (PDCAAS) (Schaafsma 2000) and the digestible indispensable amino acid score (DIAAS) (UN FAO 2013):


Higher PDCAAS values are associated with higher-quality protein. “Limiting amino acid” and “dietary indispensable amino acid” both refer to the AA of interest. The main difference between the scoring methods is that PDCAAS measures AA digestibility through fecal sample testing, while DIAAS measures AA digestibility through ileal digesta analysis. Animal proteins typically have high AA scores (>95), while non-animal proteins often have lower scores, although some exceptions exist, such as soy, myco-, and potato proteins. GFI’s plant protein primer includes the PDCAAS values (represented as fractions of 0–1.0, not as percentages) for proteins from 20 crops, some of which are reflected in Table 1 above. Herreman et al. 2020 includes DIAAS values for various plant and animal sources, including scores for optimized plant protein blends. Strategies such as combining proteins from multiple crops, optimizing crop varietals through breeding, hydrolyzing the protein, and removing antinutrients and other impurities through processing can enhance AA scores (Sa et al. 2019). Other contaminants in plant protein powders that can affect functional properties include pigments—such as chlorophyll, which produces a green color that’s difficult to remove—and off-flavors, such as those produced via lipoxygenase-catalyzed peroxidation of unsaturated fatty acids. These impurities should be masked, reduced, or eliminated to reach parity with animal-based products.
Table 2: Amino acid (AA) content of various dietary protein sources and recommended daily intake for human adults from the WHO. AA content values of crops greater than or equal to those in egg are bolded.
| Amino acid(s) | Amino acid content (g per 100 g of raw material | Recommended daily intake (g per 70 kg body weight) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Oat | Pea | Lupin | Potato | Wheat | Soy | Egg | Milk | ||
| Histidine (H) | 0.9 | 1.6 | 1.2 | 1.4 | 1.4 | 1.5 | 0.9 | 1.9 | 0.7 |
| Isoleucine (I) | 1.3 | 2.3 | 1.5 | 3.1 | 2.0 | 1.9 | 1.6 | 2.9 | 1.4 |
| Leucine (L) | 3.8 | 5.7 | 3.2 | 6.7 | 5.0 | 5.0 | 3.6 | 7.0 | 2.7 |
| Lysine (K) | 1.3 | 4.7 | 2.1 | 4.8 | 1.1 | 3.4 | 2.7 | 5.9 | 2.1 |
| Methionine (M) | 0.1 | 0.3 | 0.2 | 1.3 | 0.7 | 0.3 | 1.4 | 2.1 | 0.7 |
| Cysteine (C) | 0.4 | 0.2 | 0.2 | 0.3 | 0.7 | 0.2 | 0.4 | 0.2 | 0.3 |
| Phenylalanine (F) | 2.7 | 3.7 | 1.8 | 4.2 | 3.7 | 3.2 | 2.3 | 3.5 | Total 1.8 |
| Tyrosine (Y) | 1.5 | 2.6 | 1.9 | 3.8 | 2.4 | 2.2 | 1.8 | 3.8 | |
| Threonine (T) | 1.5 | 2.5 | 1.6 | 4.1 | 1.8 | 2.3 | 2.0 | 3.5 | 1.1 |
| Valine (V) | 2.0 | 2.7 | 1.4 | 3.7 | 2.3 | 2.2 | 2.0 | 3.6 | 1.8 |
Fibrous and globular proteins
As noted, the lower digestibility of plant proteins is partly due to the contrasting structures of plant and animal proteins. In addition to AA sequence and composition, protein function depends on other structural features, such as total molecular weight and configuration. Physicochemical features, including hydrophobicity, charge, and reactive groups—free thiols or thioethers from sulfur-containing amino acids, for example—play an essential role in configuration. They trigger various mechanisms including hydrophobic, electrostatic, and covalent interactions to form secondary protein structures, tertiary protein structures, and quaternary protein structures, all of which affect functionality. For instance, plant proteins generally have more β-sheet secondary structures and fewer α-helices than animal proteins. β-sheet structures have shown resistance to digestion, which is partly why plant proteins generally have lower PDCAAS and DIAAS values than animal proteins (Nguyen et al. 2015; Carbonaro et al. 2012). Beyond digestibility, all other protein functions are affected by structure.
The two main types of proteins that are especially important when comparing animal and plant proteins are fibrous and globular proteins (Figure 3). Fibrous proteins are typically structural and protective, forming filamentous structures, such as connective tissue and muscle fiber. They have repetitive AA sequences, are mostly water-insoluble, and often interact through covalent or noncovalent crosslinking. While fibrousness is primarily associated with animal proteins, some non-animal proteins, such as glutenin and mycoprotein, have fibrous structures (McClements and Grossmann 2021).
On the other hand, globular proteins can act as functional and structural proteins, have irregular AA sequences, and are more water-soluble. Their structures are more tightly packed and sensitive to environmental conditions (such as pH and temperature) than fibrous protein structures. Most plant proteins are globular and require chemical, mechanical, or biological processing to mimic the fibrous structure of animal proteins. During restructuring, globular proteins are denatured, unfolded, aligned, and crosslinked through interactions like hydrophobic effects, van der Waals, hydrogen bonding, electrostatic effects, and disulfide bonding. The deep dives for ingredient optimization and end product formulation and manufacturing detail globular protein enrichment and texturization. Still, knowledge of native protein structure is essential for optimizing end product texture.
Animal-derived proteins
Animal skeletal muscle is attached to bones by tendons and has a complex, hierarchical structure composed of elongated fibrous cells. The aggregation and bundling of fibrous animal proteins provide unique properties challenging to reproduce with non-animal proteins. Bomkamp et al. 2021 and Listrat et al. 2016 provide more details on fibrous protein bundling and animal-muscle tissue structure. Here, we briefly summarize the individual proteins that make up animal-based products.
Generally, about 10% of skeletal muscle is composed of connective and fat tissues. Collagen is the primary structural protein in animal connective tissues; it is a triple helix of intertwined polypeptides that forms an elongated, fibrous protein. The other 90% is mostly muscle fibers and contains proteins—chiefly, myosin, hemoglobin, and sarcoplasm proteins, including myoglobin and actin. Myosin’s elongated structure forms macromolecular filaments through interactions of multiple subunits. Actin is interesting, in that it polymerizes with itself to form thin microfilaments that interact with myosin and thus contribute to skeletal muscle’s filamentous nature, even though actin is a globular protein. Hemoglobin and myoglobin are globular but serve the functional purpose of transporting oxygen. Hemoglobin operates in blood, while myoglobin is found in skeletal muscle. Heme proteins are responsible for the red and brown colors in meat because they change colors according to their iron atom’s oxidation state. At ambient conditions, animal meat is red because the iron atoms in heme proteins are in an oxidized state (Fe2+) and bound to oxygen molecules (O2). The meat turns brown upon cooking or prolonged storage as the iron atoms release oxygen and lose an electron (Fe3+). This color change has proved difficult to replicate with non-animal proteins, although Impossible Burger’s soy leghemoglobin, derived through fermentation technology, demonstrates an innovative solution.
Animal-based dairy products are similarly challenging to replicate with plant proteins because of their predominant proteins’ remarkable structures and functions. Here, we briefly discuss the characteristics of the major protein in dairy, casein. Casein has a high DIAAS value of 117 and a balanced AA profile (Herreman et al. 2020). Casein also has a unique random-coil structure that is neither fibrous nor globular but is flexible, anionic, and strongly amphiphilic (it has a balance of polar and nonpolar regions) (Figure 3). As a result, it forms micelles in an aqueous solution that divalent ions, such as calcium, can further crosslink. In milk, caseinate (casein’s ionic form) and its micelles emulsify and gel around fat droplets, forming a colloidal solution. Casein’s electrostatic, amphiphilic, and structural features have made the food ingredient attractive beyond its natural application in milk—for cheese, yogurt, ice cream, meat, pasta, and baked goods. Unfortunately, no known plant proteins resemble casein structurally, which makes replicating its textural, nutritional, matrix-forming, fat- and water-binding, and stabilizing attributes challenging. Some companies are working to produce casein without animals. Nobell Foods, for example, applies molecular farming to produce casein from soybeans, and a number of startups are producing casein through microbial fermentation. The crop breeding section of this deep dive describes molecular farming in more detail.
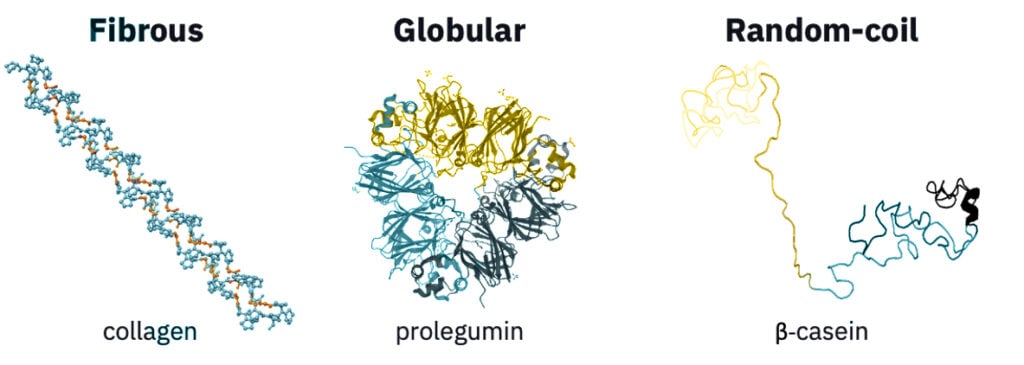
Source: NIH protein database for collagen (1K6F) and pea prolegumin (3KSC) structures and Sun et al. 2021 for β-casein structure.
Plant-derived proteins
While animal protein structures and functions differ significantly from those of plant proteins, native plant proteins have interesting properties that can create similar end products. Plant proteins have varied abilities that depend on their crop sources, even within a plant family. For example, while legumes are often bulked together as nitrogen-fixing crops, notable diversity is observable among their protein features—mung bean and chickpea proteins have superior gelling properties compared with lentil, lupin, and faba bean proteins (Kyriakopoulou et al. 2021). Just as animal proteins are combinations of collagen, myosin, actin, myoglobin, etc., crop proteins are heterogeneous mixtures of proteins with distinct structures and functionalities. The crop species and cultivar, growing and harvesting conditions, and downstream processes affect the types and distributions of these proteins.
Globular plant proteins are divided into four categories defined by their aqueous solubility: globulins (soluble in salt water), albumins (soluble in water), prolamins (soluble in alcohol or alcohol-water mixtures), and glutelins (soluble in acidic or alkaline water). Protein solubility in water depends on AA composition and sequence, molecular configuration and size, , and physicochemical characteristics, such as surface polarity, net charge, and reactive groups. Solubility affects other functionalities, such as a protein’s thermal stability, emulsifying and foaming properties, and gelling abilities. As a result, globulin, albumin, prolamin, and glutelin protein fractions from the same crop source have distinct functionalities. For instance, globulin and albumin from a Ginkgo seed protein isolate demonstrate significantly different properties. For instance, the albumin fraction has better emulsifying and oil-adsorption abilities (Deng et al. 2011). Pea albumin proteins form firmer gels than pea globulin proteins (Kornet et al. 2021). For Chinese quince seed, glutelin proteins demonstrate better thermostability and oil-adsorption capacity than the albumin fraction (Deng et al. 2020). Understanding the categories of globular plant proteins sheds light on approaches to optimizing their functionalities:
- Globulins are generally insoluble in pure water but dissolve in highly concentrated salt solutions. They have higher molecular weights than albumins. 7S globulins and 11S globulins are common plant proteins primarily found in legume-crop seeds. Note that 7S and 11S refer to the sedimentation coefficients that reflect a protein’s molecular weight and viscosity. These globulin families are separable upon fractionation. Even within a protein family, protein fractions have distinct properties: 7S globulins are usually glycosylated and have fewer cysteine residues, so they cannot easily form disulfide bridges. 11S globulins, however, have two conserved disulfide bridges (Tandag-Silvas et al. 2011). Common globulins in crops used for alternative protein products include cruciferin (12S), legumin, edestin or glycinin (11S), and vicilin or β-conglycinin (7-8S). Globulins tend to have relatively high levels of basic AAs (lysine and arginine) and low levels of glutamine, glutamic acid, and proline (Cornell 2012).
- Albumins are soluble in pure water and saline solutions at low concentration. They have generally low molecular weights (<50 kDa) and so exhibit lower sedimentation coefficients than globulins. They are also rich in cysteine residues and have few hydrophobic groups compared with less-water-soluble proteins. 2S albumins exist in many crop seeds and include hypervariable regions in their AA sequences that may induce allergic reactions (Souza 2020). Albumins tend to have higher cysteine content and lower glutamine/glutamic acid and proline content than prolamins and glutenins.
- Prolamins are soluble in alcohol or alcohol-water solutions and have high proline and glutamine AA content. Gliadin, zein, avenin, and kafirin are prolamin proteins primarily found in cereal crops. Gliadin and other Triticeae (e.g., wheat, barley, rye) prolamins are the primary triggers for celiac disease.
Glutelins form a low-solubility subclass of prolamins that require acidic or basic water for solubility and are dubbed acidic or basic glutelins accordingly. Both prolamins and glutelins tend to have higher glutamine and proline levels and lower cysteine content than globulins and albumins.
Crop protein content
Table 3: Properties of the major proteins in crop sources
| Protein content (%) | Molecular weight (kDa) | Isoelectric point (pI) | Melting temp. (Tm) (ºC) | Comments | |
|---|---|---|---|---|---|
| Soy | |||||
| Glycinin (11S) | 36 to 51 | 300 to 380 | 4.5 | 93 | Trimer, globulin |
| β-conglycinin (7S) | 17 to 24 | 150 to 200 | 5 | 80 | Hexamer, globulin |
| Pea | Multimers | ||||
| Legumin (11S) | 55 to 80 | 360 | 4.5 | 75 to 79 | Globulin |
| Vicilin (7S) | 150 | Globulin | |||
| Convicilin | 280 | Globulin | |||
| Albumin (2S) | 18 to 25 | 50 | 6.0 | 110 | Albumin |
| Corn (maize) zein | Hydrophobic | ||||
| α-zein | 75 to 85 | 19 to 24 | 6.4 | 89 | Packed helices |
| β-zein | 10 to 15 | 14 to 15 | |||
| γ-zein | 5 to 10 | 16 to 27 | |||
| Canola | |||||
| Globulins | 60 | 14 to 59 | 4.5 | 84 to 102 | Mainly cruciferin (12S) |
| Albumins | 20 | Mainly napin (2S) | |||
| Glutelins | 15 to 20 | ||||
| Prolamins | 2 to 5 |
Cereals mostly contain prolamins and glutelins with the notable exceptions of oat and rice crops, which are composed chiefly of 11S globulin proteins. On the other hand, legumes more commonly contain predominantly globulins (Tandang-Silvas et al. 2011). While crop proteins are often described as a single protein (e.g., “soy protein isolate”), they are heterogeneous mixtures of different types of proteins. For example, soy (Glycine max) protein isolate is a blend of two primary globulin proteins, glycinin (11S) and β-conglycinin (7S) (McClements and Grossmann 2021). The AA sequences and structures of glycinin and β-conglycinin are different, which creates distinctions in their solubilities and other properties (Khatib et al. 2006). Due to its free cysteine residues, glycinin can form disulfides that contribute to the formation of stronger gels than β-conglycinin, which does not have available cysteines (Renkema et al. 2001; Choi et al. 2006; Tandang-Silvas et al. 2011). The distribution of these protein fractions in flour depends on crop variety (Yaklich 2001; Khatib et al. 2006) and protein processing conditions (Rickert et al. 2006), so different crops and enrichment methods may produce final products with significantly distinctive properties, despite having the same botanical origin. For soybean, protein fractions combine all the essential amino acids necessary for human nutrition and desirable properties such as emulsification and gelation. The functionalities described in GFI’s plant protein primer convey the attributes of whole protein isolates. An understanding of plant proteins can be furthered by connecting a protein product’s functionality to the ratios, sequences, and structures of its protein fractions. Table 3 (modified from McClements and Grossmann 2021) displays the protein fractions in crops along with some of their properties. More details are discussed below for select crops.
Pea (Pisum sativum) protein also mainly contains globulin proteins, specifically legumin (11S) and vicilin (7S), as well as convicilin (7-8S) and albumin (2S). These fractions have distinct properties—vicilin has demonstrated emulsification and gelation properties superior to those of legumin (Barac et al. 2015). Additionally, although 2S albumin has desirable levels of sulfur-containing AAs (the limiting AAs in pea protein), it is also reported to have antinutritional properties that cause low digestibility and induce allergic reactions (Malley et al. 1975; Souza 2020).
Wheat (Triticum) gluten is known for its binding and dough-forming capacity, viscosity, and nutritional quality (Delcour et al. 2012). Gluten is composed of two main components, gliadins and glutenins, where the relative amounts and compositions of these proteins determine gluten-dough viscosity and elasticity (Barak et al. 2012). Glutenin favorably polymerizes to form intermolecular disulfide bonds, which are crucial for gluten-dough elasticity. On the other hand, gliadins contribute more to the viscosity of the network, mostly form intramolecular disulfide bonds, and must be heated to initiate gliadin-glutenin crosslinking. As a result, balancing the toughness, adhesiveness, and cohesiveness of gluten products depends on the ratios of its protein fractions.
Maize (Zea mays) protein, or corn zein, is composed of fractions simply dubbed α, β, γ, and δ (Mattice et al. 2020). Zein has a high proportion of nonpolar AAs, making it water-insoluble and imparting unique self-assembly properties. As a result, zein forms viscoelastic networks in water that are flexible. Unlike gluten, these networks are primarily driven by noncovalent interactions since the main constituent, the α fraction, does not contain many free thiols. Commercial zein is currently optimized by creating a protein product composed mainly of the α fraction. Although the β fraction accounts for 10% of corn zein, it is removed, chiefly during purification, as it is unstable, precipitates, and coagulates (Lorenzo et al. 2018).
Rapeseed, or canola (Brassica napus), consists mainly of cruciferin (12S globulin) and napin (1.7-2S albumin). Cruciferin forms stronger gels and has better emulsification stability than napin, while napin has better foaming properties than cruciferin (Akbari et al. 2015). Cruciferin creates such strong heat-set gels under alkaline conditions that Merit Functional Foods uses a cruciferin-rich, non-GMO canola protein as a binder (e.g., methylcellulose) replacement. Thus, complete protein fraction isolation is not necessary to take advantage of a protein’s traits—efforts to optimize the ratio of protein fractions include finding and developing novel crop proteins.
For example, while vicilin is dominant in pea protein isolate, faba (Vicia faba) bean contains more legumin than vicilin, demonstrating that finer tuning of crop protein properties is possible (Robinson et al. 2019). Note that similar protein fractions from different crop sources can have significantly varying properties. One study demonstrated that 7S globulins from lupin (Lupinus luteus) had a 10K higher denaturation temperature than the 7S globulins from soy (Sousa et al. 1995). Another study found functional differences among 7S and 11S globulins from soy, pea, faba bean, cowpea, and French bean (Kimura et al. 2008). Even between cultivars, differences in structure can lead to distinct functionalities. Five wild soybeans were found to have microheterogeneity in their glycinin subunits, resulting in unique gelling properties of glycinins isolated from the different cultivars (Nakamura et al. 1984). These many possible variations of protein structures and their effects on functionalities can be overwhelming to keep track of, which is why analytical services that evaluate batch-to-batch ingredient variations and open-access databases that focus on protein structure-function relations would be beneficial. Moreover, pairing these databases with machine learning, such as NotCo’s Giuseppe algorithm, can propel the creation of innovative plant-protein products.
RuBisCO (ribulose-1,5-bisphosphate carboxylase oxygenase, ~550 kDa) has recently gained significant attention for its potential use in food (Di Stefano et al. 2018). RuBisCO is the most abundant protein on Earth due to its significant role in photosynthesis. It is derivable from any photosynthetic tissue or organism, including the green leaves of any plant crop, as well as a host of nonconventional sources, such as algae, Lemna (duckweed), and cyanobacteria. It is most soluble in alkaline pH (Lamsal et al. 2007), and digested RuBisCO peptides from chia seed (Salvia hispanica L.) were mainly found in the glutelin fraction (Grancieri et al. 2019). In addition to its abundance, RuBisCO has a complete AA profile (Zengin et al. 2012), forms heat-induced gels at lower concentrations than other plant proteins (Martin et al. 2014), and functions as a good foaming and emulsifying agent (Barbeau et al. 1988). Unfortunately, it also contains high levels of off-flavor compounds and chlorophyll pigment that imparts a green color and undesirable flavor that are difficult to mask or remove. Additionally, retaining the techno-functional attributes of RuBisCO after typical commercial-scale enrichment has proved difficult. NIZO developed a scalable extraction process for RuBisCO purification that yields a colorless protein isolate. Plantable Foods claims a proprietary organic cold-press extraction process that yields a white RuBisCO product from Lemna. The company states that Lemna produces more digestible RuBisCO than pea, soy, or algae.
These few examples make clear that plant proteins are incredibly diverse. Yet many more raw material sources remain underutilized. Some university labs, established companies, and startups are focused on unleashing the power of novel crop sources, such as algae (Kazir et al. 2019; Trophic LLC), chickpea (InnovoPro, NuCicer), Pongamia tree (TerViva), and quinoa (NorQuin with Ingredion).
Beyond these, the opportunities to investigate underexplored crops are seemingly endless, as illustrated by GFI APAC’s Asian Cropportunities report and GFI Brazil’s 2021 research funding program, which focus on crops endemic to Asia and Brazil, respectively. Researching novel crop sources could aid in creating innovative products for many different cultures and thereby increase agricultural biodiversity and promote use of indigenous crops. For example, an African research team found that a protein composite from lima beans and African oil bean seeds, both indigenous to Africa, offered a balanced essential AA composition (Arueya et al. 2017). Successful application of such crops requires further characterization of the structures and functions of their proteins.
Some work has been done to characterize changes in proteins due to environmental conditions and agricultural practices. For instance, agricultural practices such as planting date, seeding rate, and row type alter the composition of proteins, oils, and fatty acids in soybeans (Bellaloui et al. 2015). Substantial room remains for information-gathering and data analysis to correlate agricultural practices and seasonal variability with compositional effects to inform when, where, and how such crops should be planted and harvested to optimize their attributes for plant-based meat end uses.
Crop genotyping and breeding for plant-based meat optimization
Besides exploring native crop sources and growth conditions, raw ingredient functionality and composition can be controlled by selecting and breeding crop cultivars. Crops have traditionally been bred for increased yield, but shifting the focus to include plant protein functionality would bolster the value of the resulting ingredients because of improved performance in specific end products (Ismali et al. 2020). Crop genetics and breeding are expansive, fast-developing research areas, and this section covers topics most relevant to crop optimization for plant-based meat applications. Other reviews offer more detailed information about crop phenomics (Yang et al. 2020; Araus et al. 2014), crop genome-wide association studies (Huang et al. 2013; Brachi et al. 2011), marker-assisted crop breeding (Xu et al. 2012), and crop breeding more generally (Varshney et al. 2006; Ahmar et al. 2020).
Identification of important genetic traits
To truly understand a crop’s protein yield, composition, sequence, structure, and function, the genes that impart these traits must be identified. The first step is understanding which traits are of interest and can be optimized. For higher-quality, functional proteins, agronomic and seed quality traits of interest include the following:
- High total protein content
- Optimal protein fraction composition
- Bland protein flavor (reduction or masking of off-flavor compounds)
- Good protein digestibility
- High micronutrient concentration
- Minimal anti- and non-nutrient factors
- Low toxicity and allergenicity
- Suitable protein functionality
- Protein fraction enrichment with minimal processing
The observable and quantifiable traits, such as those listed above, are referred to as the plant’s phenotype. They result from the crop’s environment and genetic information. To identify genetic information responsible for a crop characteristic, crops and seeds are first phenotyped—examined on the basis of trait-expression level. For example, if high seed-protein content is a desired trait, seed protein is extracted from many crop cultivars’ seeds, and then the protein concentrations are calculated and compared. This can be a time- and resource-intensive procedure, but methods such as high-throughput phenotyping have improved the process (Yang et al. 2020).
Finding biomaterials with desired traits and genes
Once phenotypic traits are observed and quantified, researchers harvest and examine the crop’s genes. The collected crop biomaterial with desired genes is called “germplasm” and can take the form of seed, stem, leaf, pollen, or any other plant cell that can be cultured into a whole plant. Germplasm genes are then extracted and associated with their phenotype. From the complete set of genes—the genotype of the germplasm—alleles specific to the phenotype are recognized by correlating trait-expression levels with observable molecular DNA markers (e.g., single nucleotide polymorphisms [SNPs] and simple sequence repeats [SSRs]). The gene sections that induce these traits—quantitative trait loci (QTLs)—can be tricky to identify if the traits are significantly influenced by environmental factors or are not highly heritable.
An example of this process postulated a set of genes that control the seed protein content of chickpeas (Upadhyaya et al. 2016). While previous researchers sequenced the genomes of many chickpea cultivars, detected molecular markers, and established high-throughput genetic analysis methods for mapping QTLs to chickpea phenotypes, most of these efforts focused on identifying genes that control crop yield or drought and salinity tolerance. This study phenotyped the seeds of chickpeas bred from two genetically different cultivars by determining their seed protein content. By utilizing the genetic information associated with these chickpea cultivars, they were able to identify six candidate genes associated with chickpea seed protein regulation. This association was conserved among the chickpeas, regardless of their otherwise diverse gene pools.
Gali et al. 2018 similarly phenotyped and genotyped bi-parental populations to identify novel QTLs. The researchers studied pea cultivars and included phenotyping from two geographies, thus taking environmental interaction into account. Using this precise genotyping, they discovered QTLs for many important traits, including seed protein concentration; seed phytate concentration; seed starch concentration; seed weight and grain yield; and concentration of seed zinc, iron, and selenium. Bi-parental crossing can have limited genomic resolution, but genome-wide association studies (GWAS) overcome some of these limitations (Huang et al. 2013; Brachi et al. 2011). Other examples of relevant phenotyping, genotyping, and QTL mapping include studies on soybeans (Seo et al. 2018; Duhnen et al. 2017), wheat (Sandhu et al. 2021), hemp (Galasso et al. 2016), and pigeonpea (Obala et al. 2019), as well as additional chickpea research (Jha 2018; Roorkiwal et al. 2020).
The USDA has open-access germplasm collections for thousands of crop accessions available through the National Germplasm Resources Laboratory. Using ~3,000 mung bean accessions from the USDA, Sandhu et al. 2020 screened Iowa field conditions for these mung beans and identified genetic markers for traits such as plant height, days to flowering, seed weight, and seed color. Similar studies should be conducted to predict and optimize more quantitative traits and properties with greater relevance to downstream functional properties for mung bean and other crops. More open-access germplasm collections, genome sequence databases, DNA marker research, and QTL maps for crops and traits relevant to protein development are necessary for the success of these studies.
Conventional (traditional) breeding
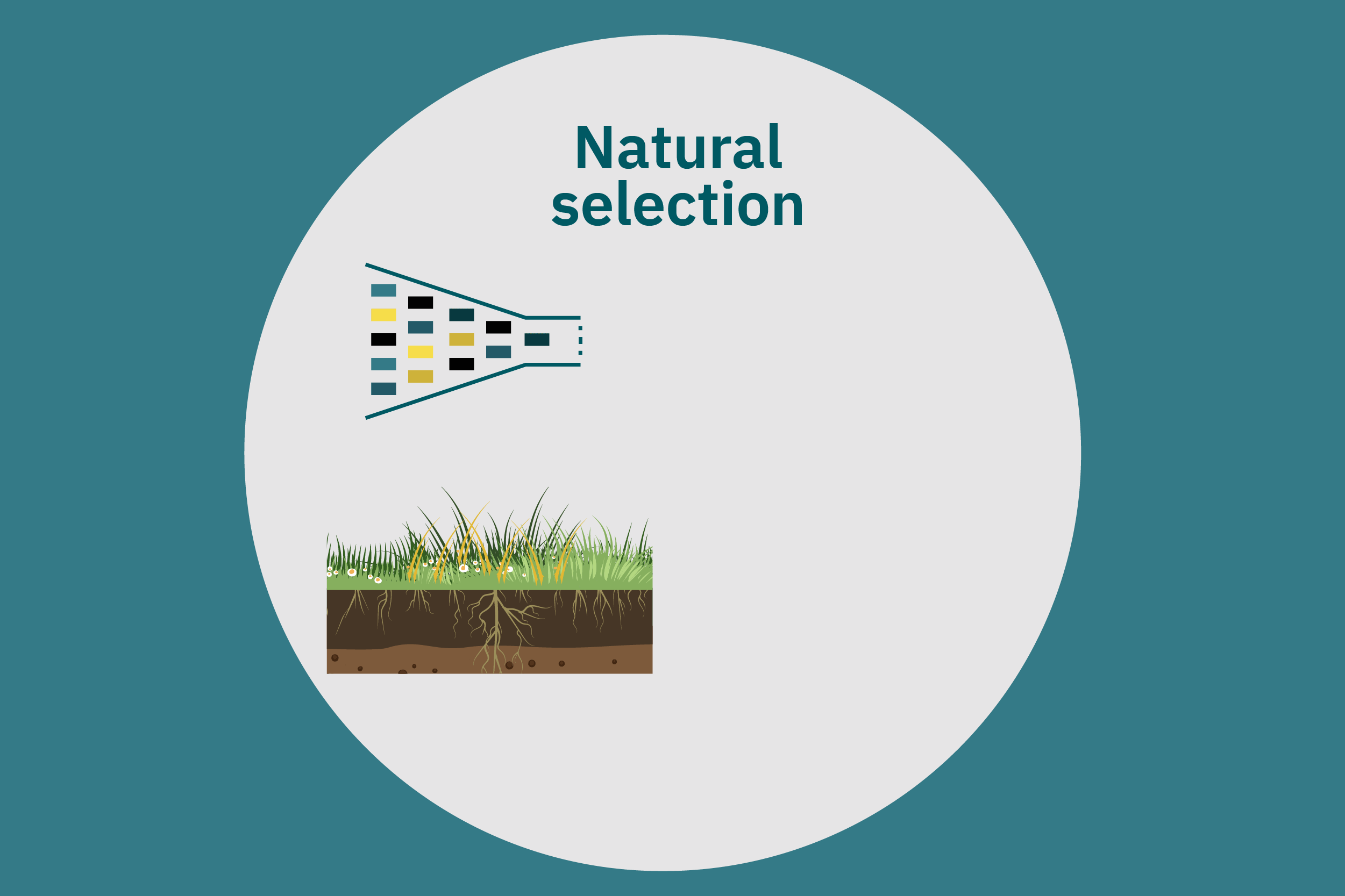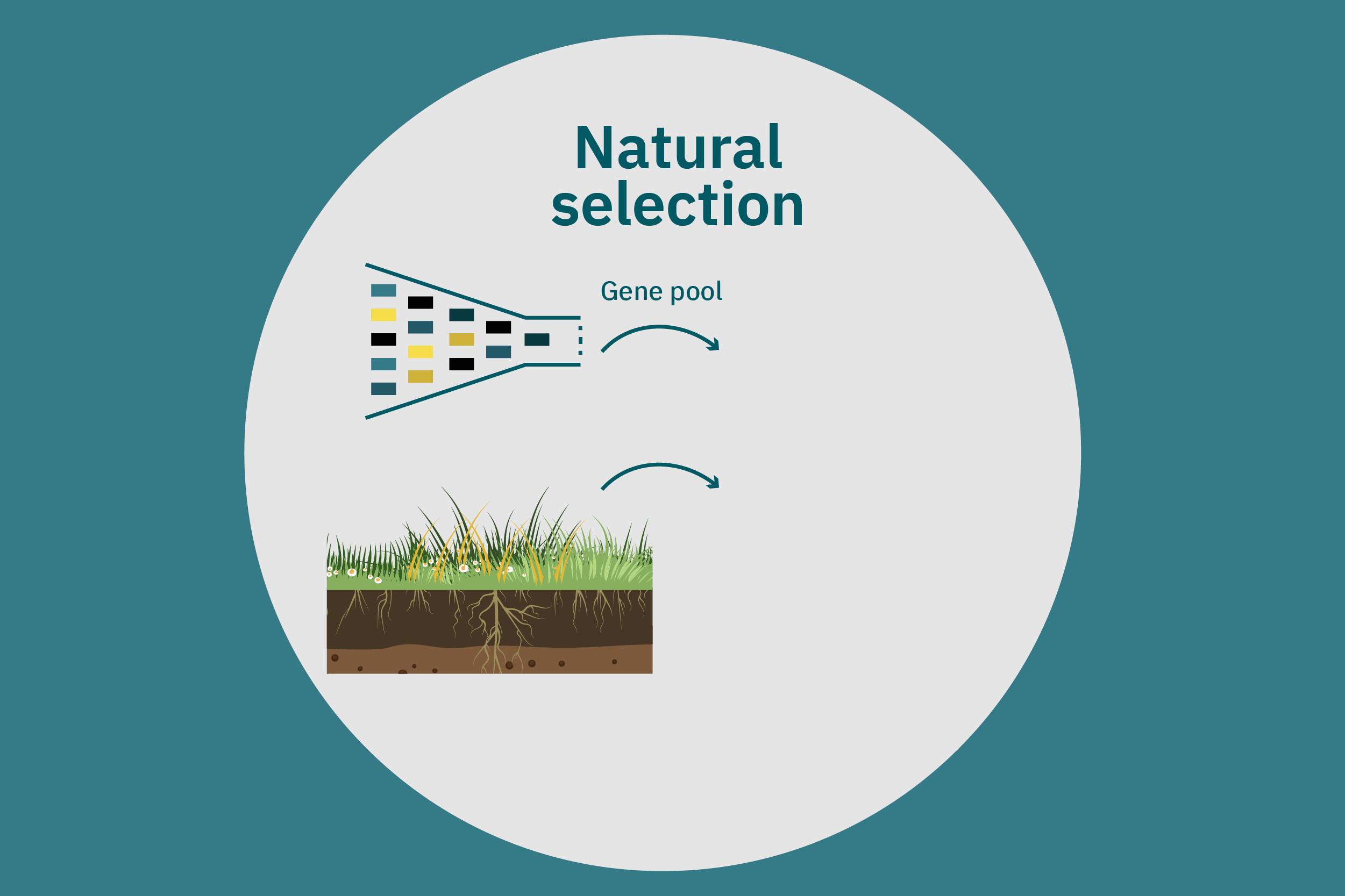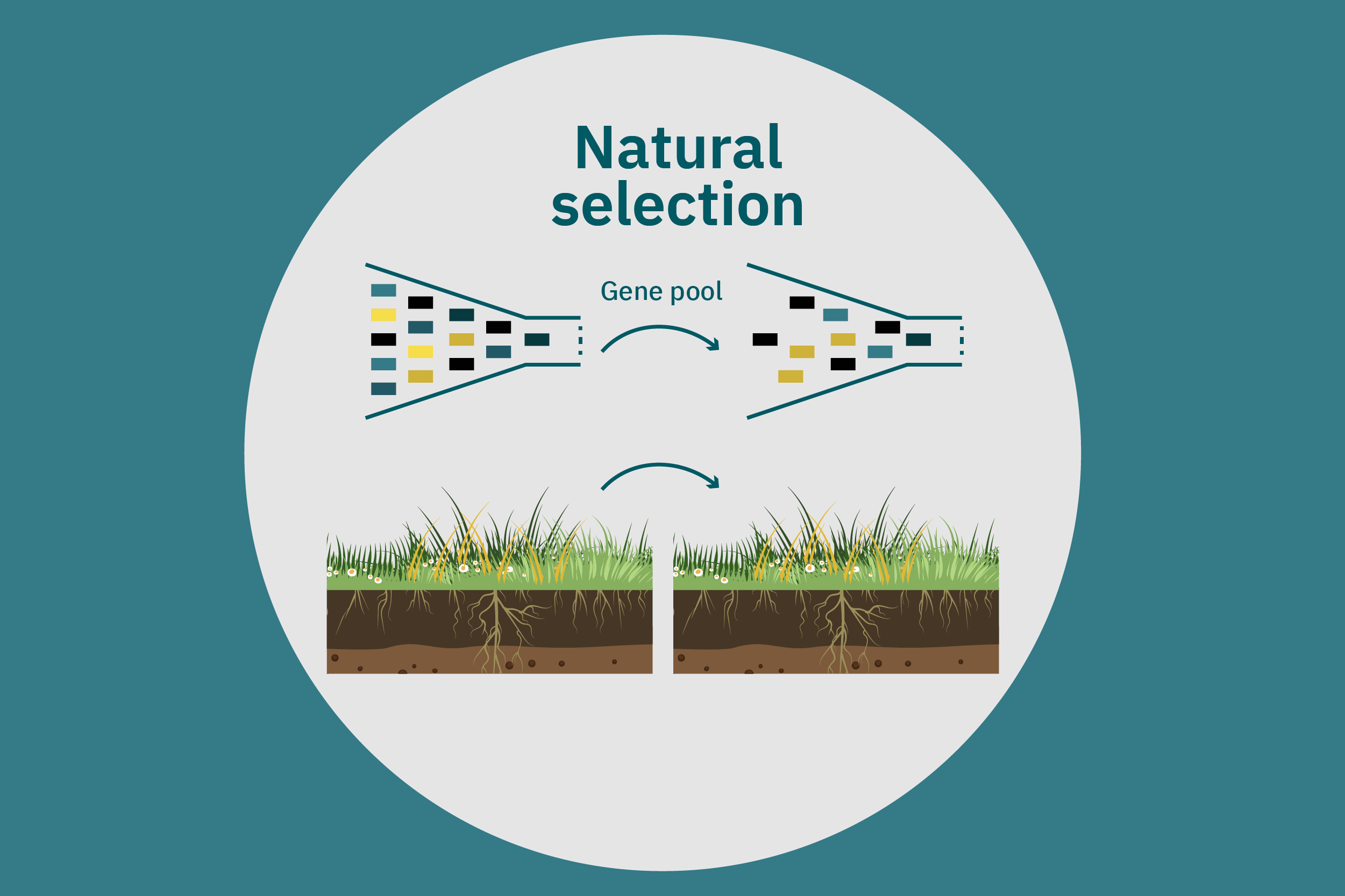
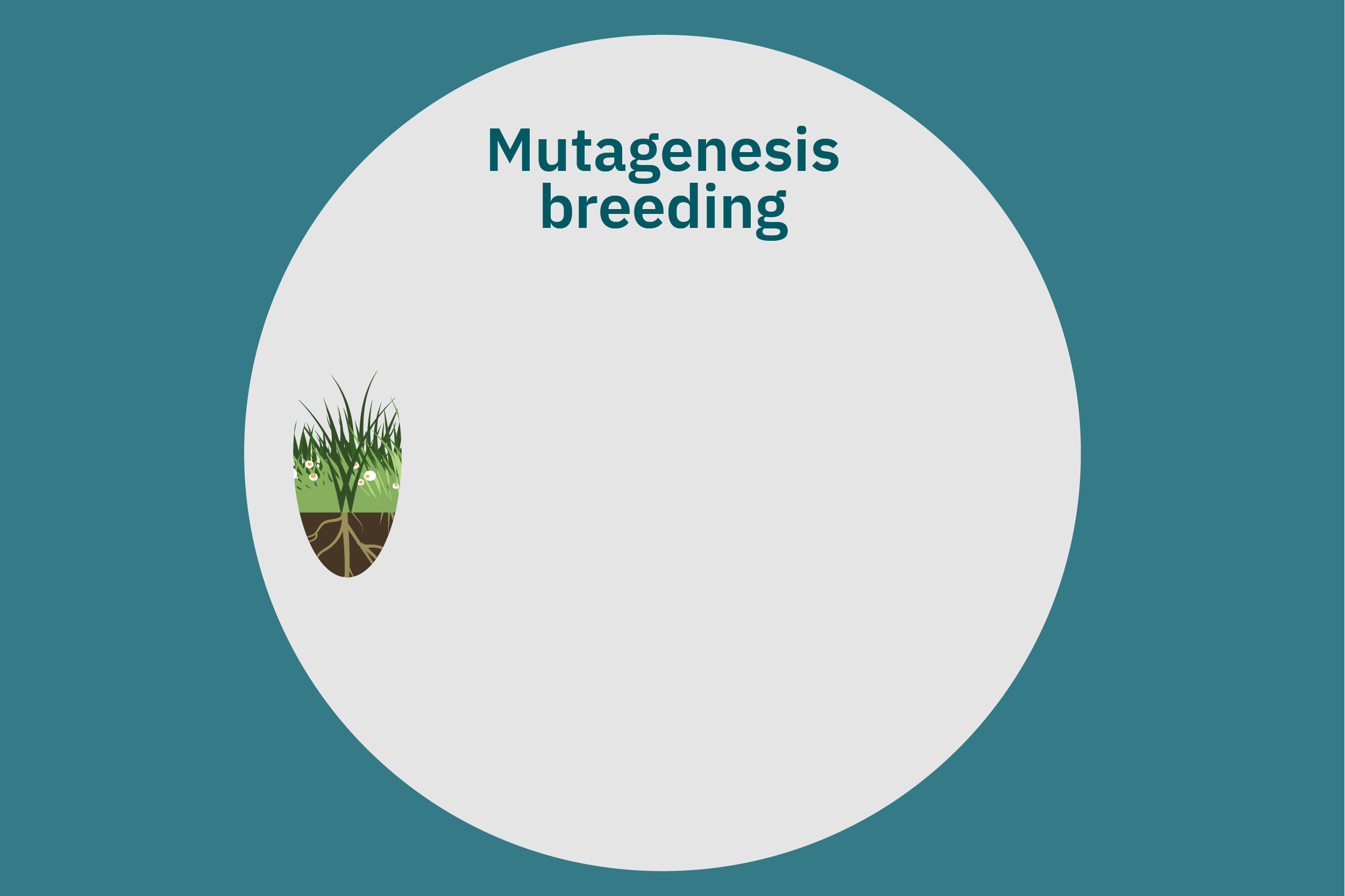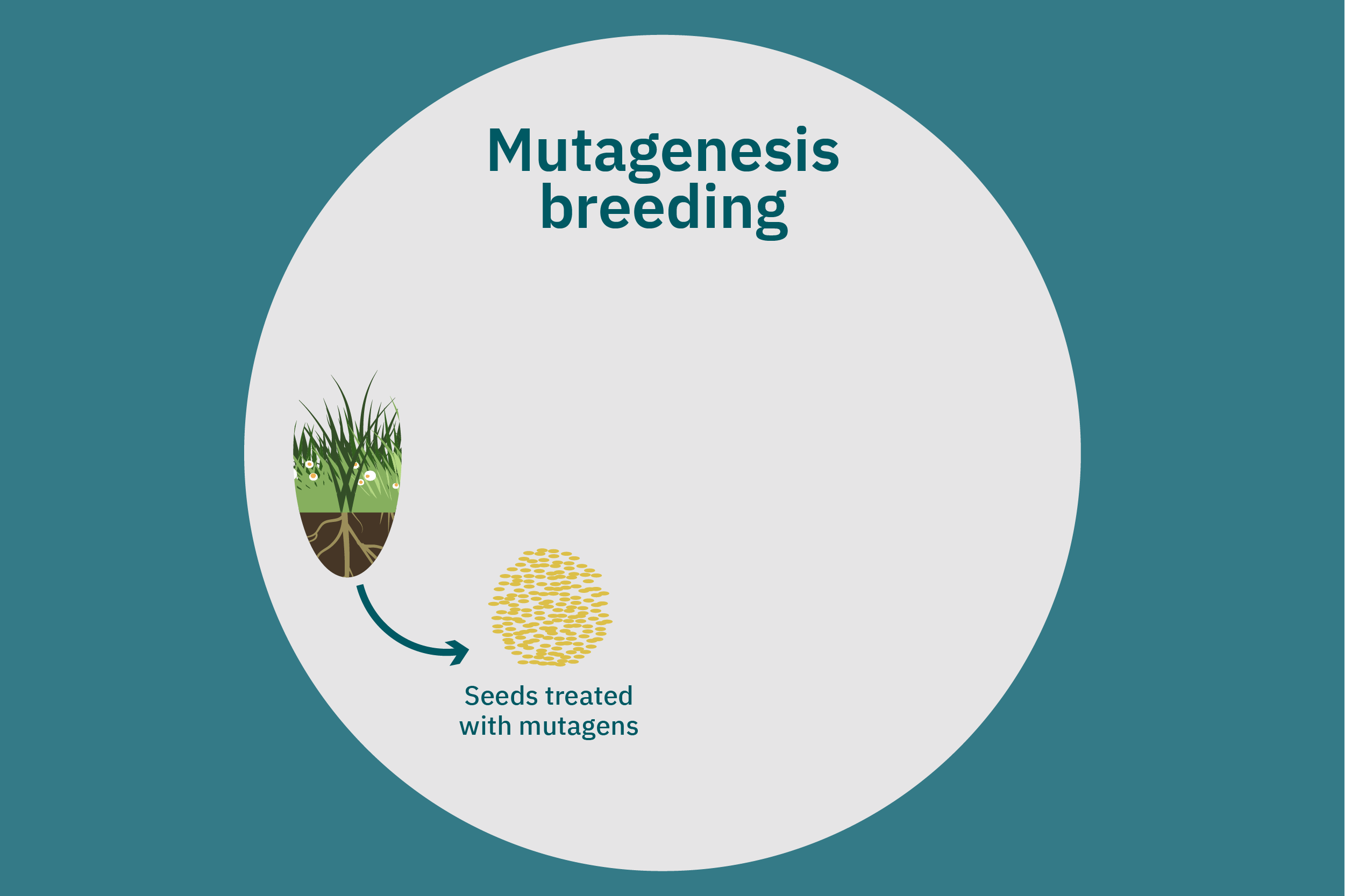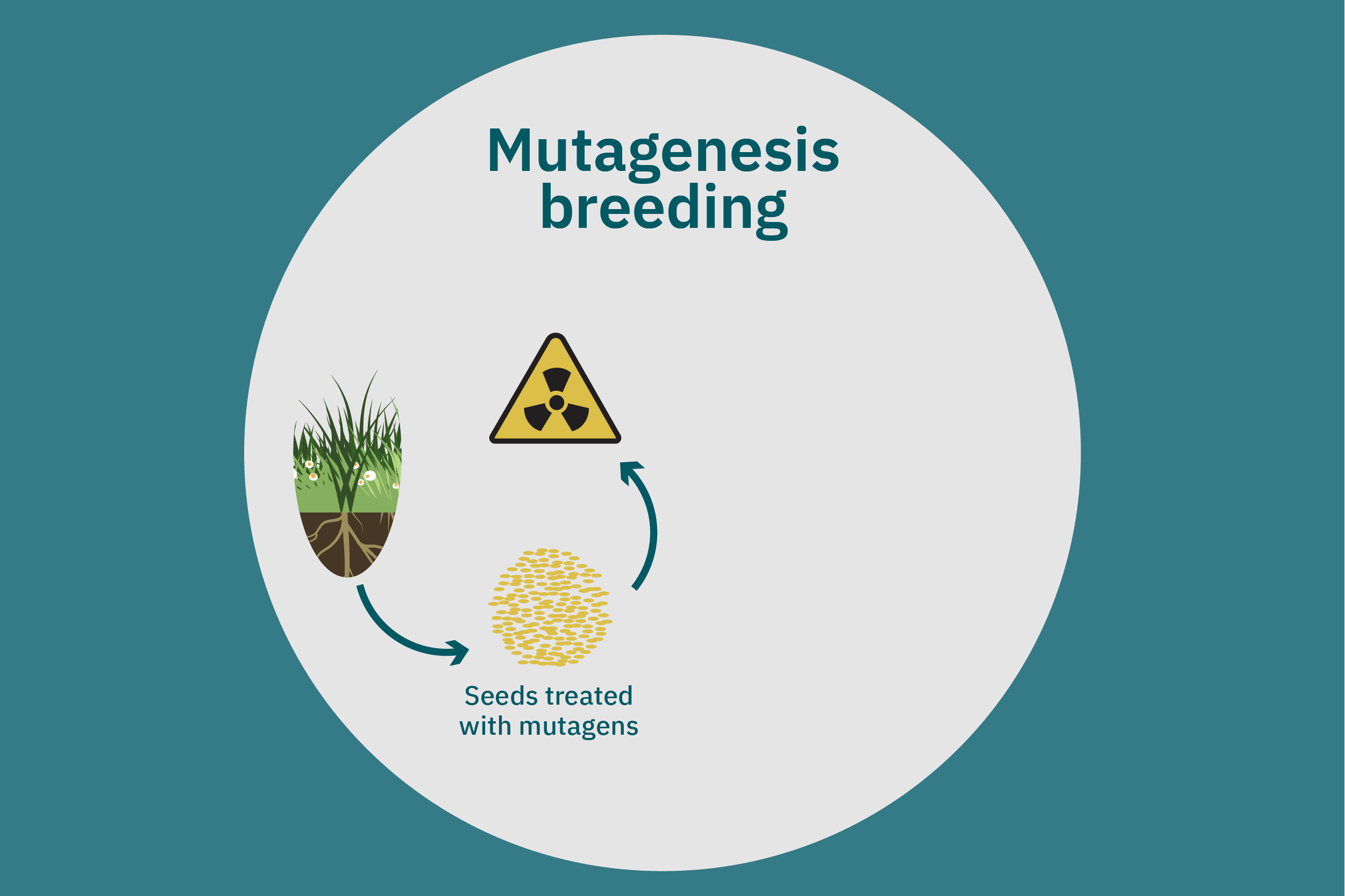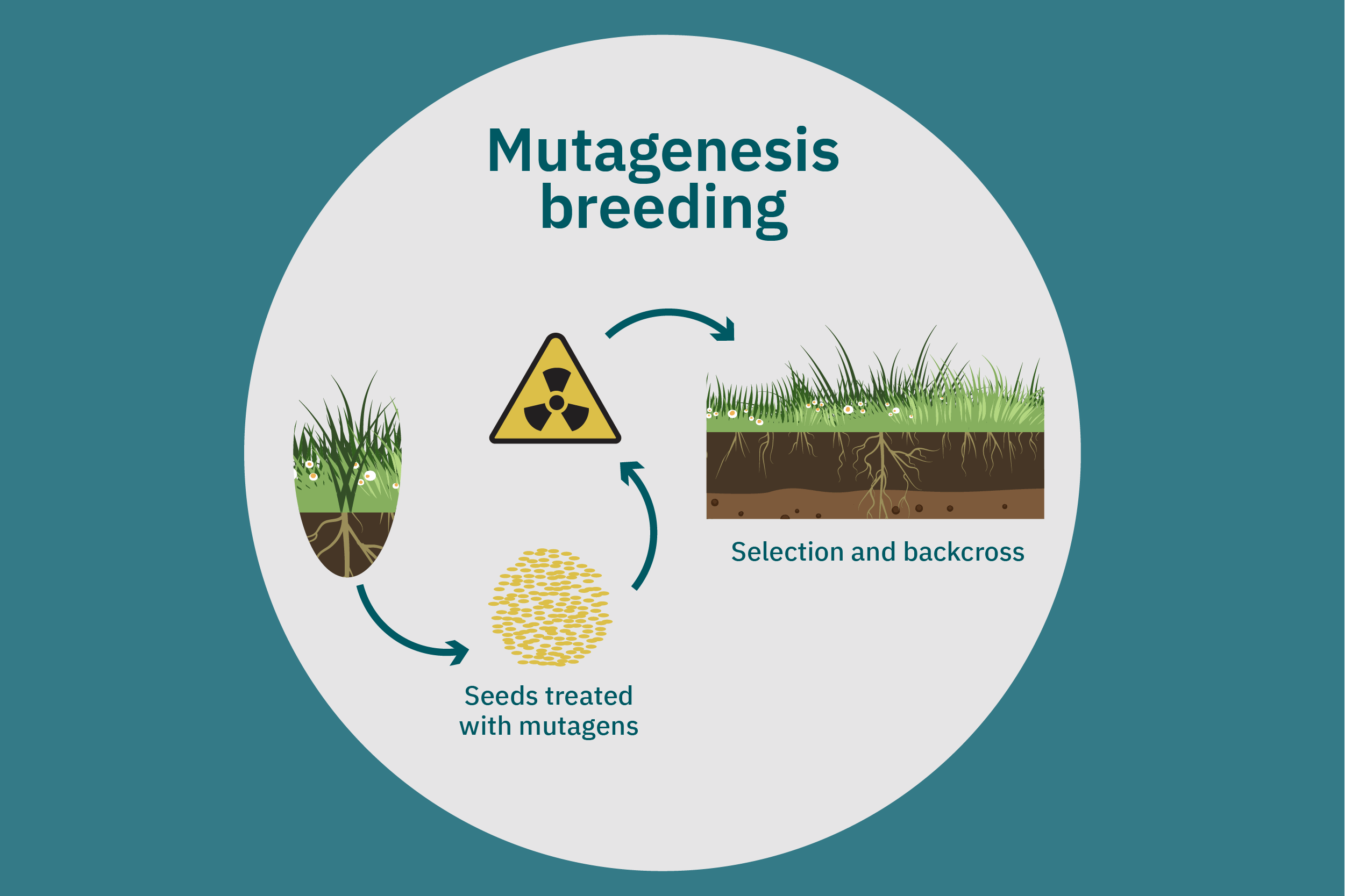
Equipped with an understanding of crop phenotype and genetics, researchers can begin cross-breeding plants with favorable genes to optimize their traits (Ahmar et al. 2020). Transferring novel genes through conventional breeding or genetic engineering enables breeders to change the phenotype of a crop. Figure 4 above illustrates various breeding and cultivar development approaches. Traditional breeding methods rely on naturally occurring genetic variations. Parent crops are cross-bred, and their progeny are selected for further breeding until the trait of interest is at the desired expression level. This type of breeding does not necessarily require genetic information. However, for more complex traits, such as biasing the expression profile of plant storage proteins to enrich the subset of the host plants’ storage proteins that perform best in plant-based meat applications, traditional breeding techniques aided by genetic characterization and high-throughput digital phenotypes may be best suited. So-called speed breeding techniques, which shorten plant reproductive cycles, can also be leveraged to accelerate strain development. Recently, to capture phenotypes that accurately reflect relevant attributes in the field, researchers used controlled lighting and growth conditions that allowed for up to six generations of crops within a single year without compromising the fidelity of each generation (Watson et al. 2018; Ghosh et al. 2018). Other techniques that bolster conventional breeding include mutation breeding and rapid generation advance, discussed in Ahmar et al. 2020.
Once promising novel protein sources for plant-based meat are identified, these crops can undergo the same concerted strain development described for improving commodity crops specifically for these applications. In addition, these novel protein sources can undergo selective breeding for the same properties—such as yield, robustness to biotic and abiotic stress, and ease of harvesting—that commodity crops have undergone over the past several decades or centuries. Improved yield and reduced losses in the field and at harvest will render these crops more attractive to farmers, which may, in turn, further decrease the cost of these proteins through both higher efficiency and economies of scale.
Examples of traditional breeding focused on enhancing protein functionality are the lupin (Lupinus mutabilis) breeding efforts outlined in Gulisano et al. 2019 and wheat breeding with simultaneous selection for high protein content and high grain yield (Michel et al. 2019). Professor Dil Thavarajah is a GFI grantee whose research focuses on genotyping and identifying molecular markers for pulse crops with alternative protein potential (Powers et al. 2021) and applying her findings to organic, traditionally bred pulses with functional traits. Companies that employ traditional, non-GMO breeding methods to advance plant protein sources include Equinom, which optimizes seed traits for soybean, cowpea, mung bean, chickpea, fava bean, and sesame; PURIS, which focuses on soy, yellow pea, and lupin; and Benson Hill, which breeds soybean and yellow pea to create an ultra-high-protein variety. For more dedicated strain development for plant-based meat applications, private companies have generated enormous databases of various germplasms’ historical performance data, genetic profiles, molecular compositions, breeding fidelity, etc., which is fed into algorithms for informing targeted breeding, editing, and engineering approaches to accelerate strain development for the target traits. For example, if the components of the biosynthetic pathways for bitter saponins have been mapped to discrete genomic loci, precision breeding or targeted pathway editing of genetically characterized strains can be used to knock these pathways down or out in as little as a single generation.
Genetic engineering and editing
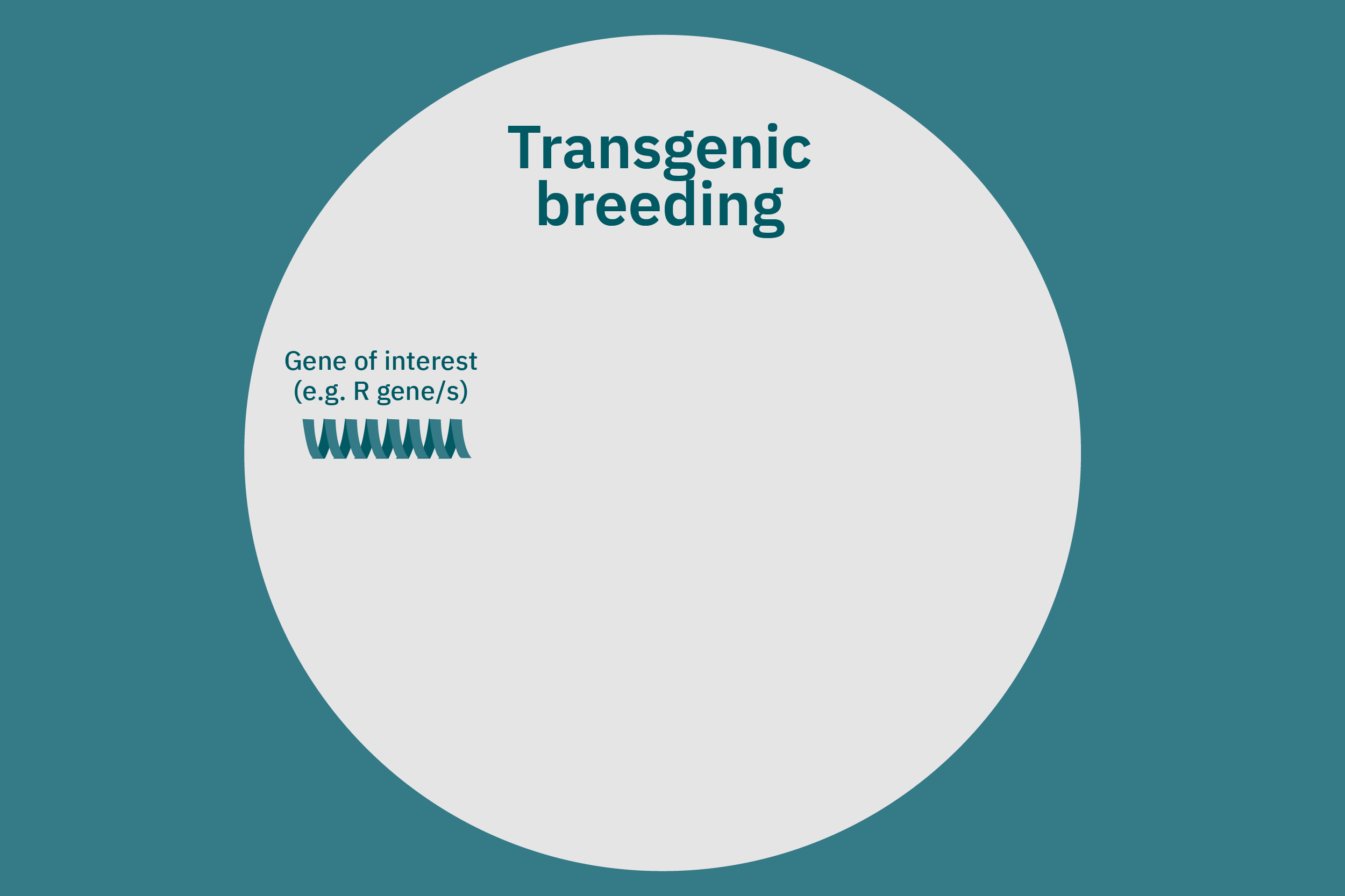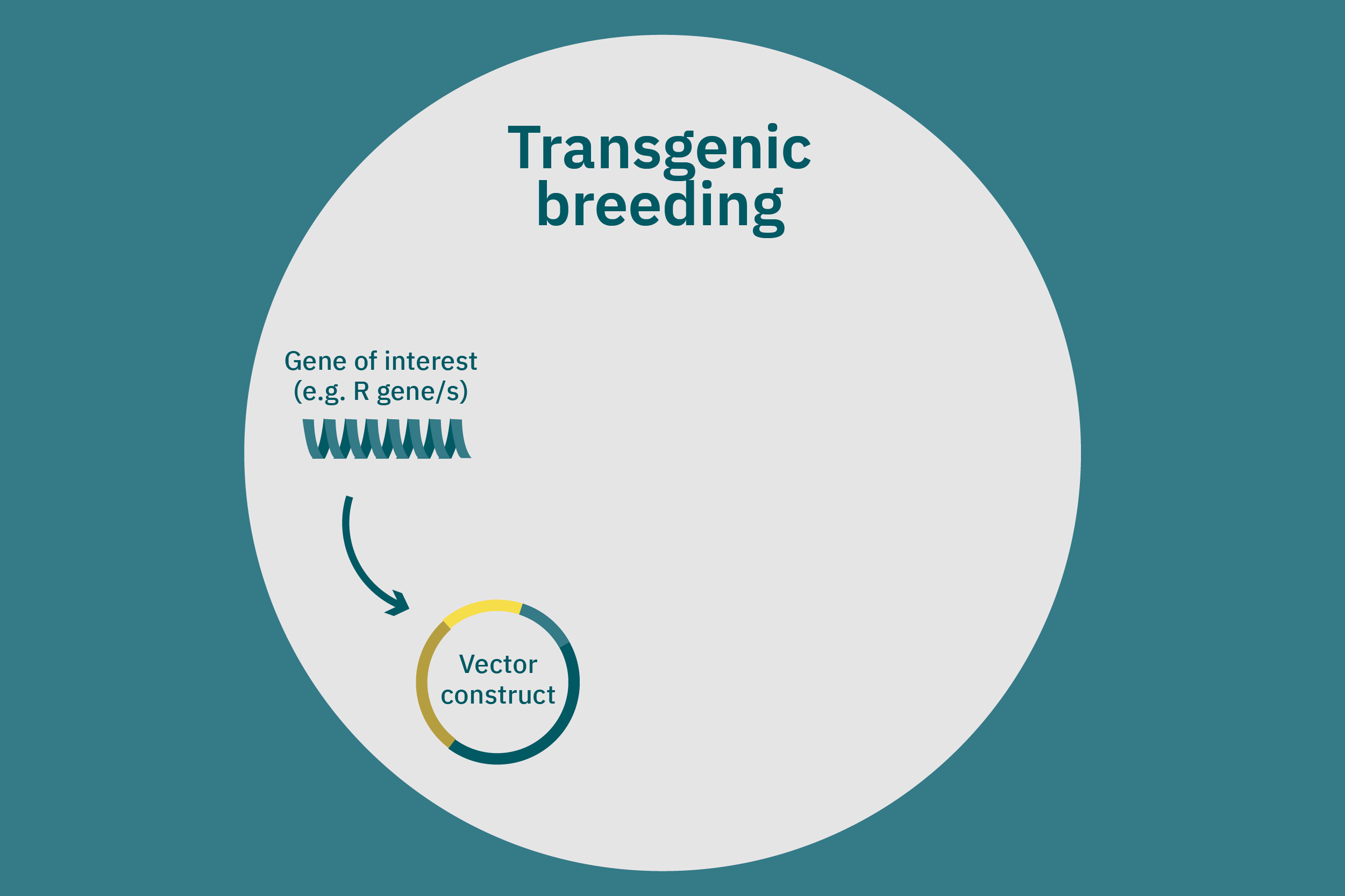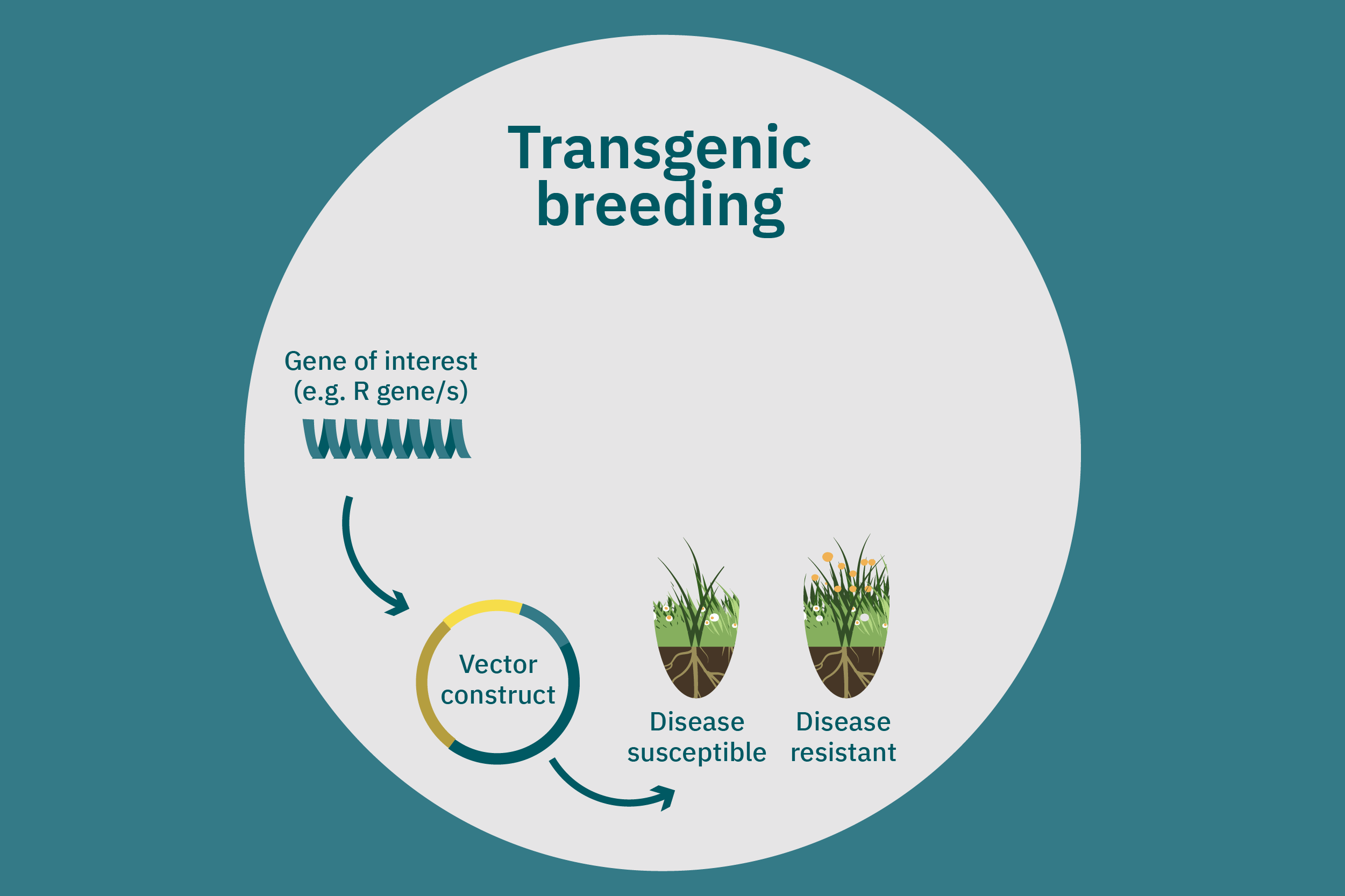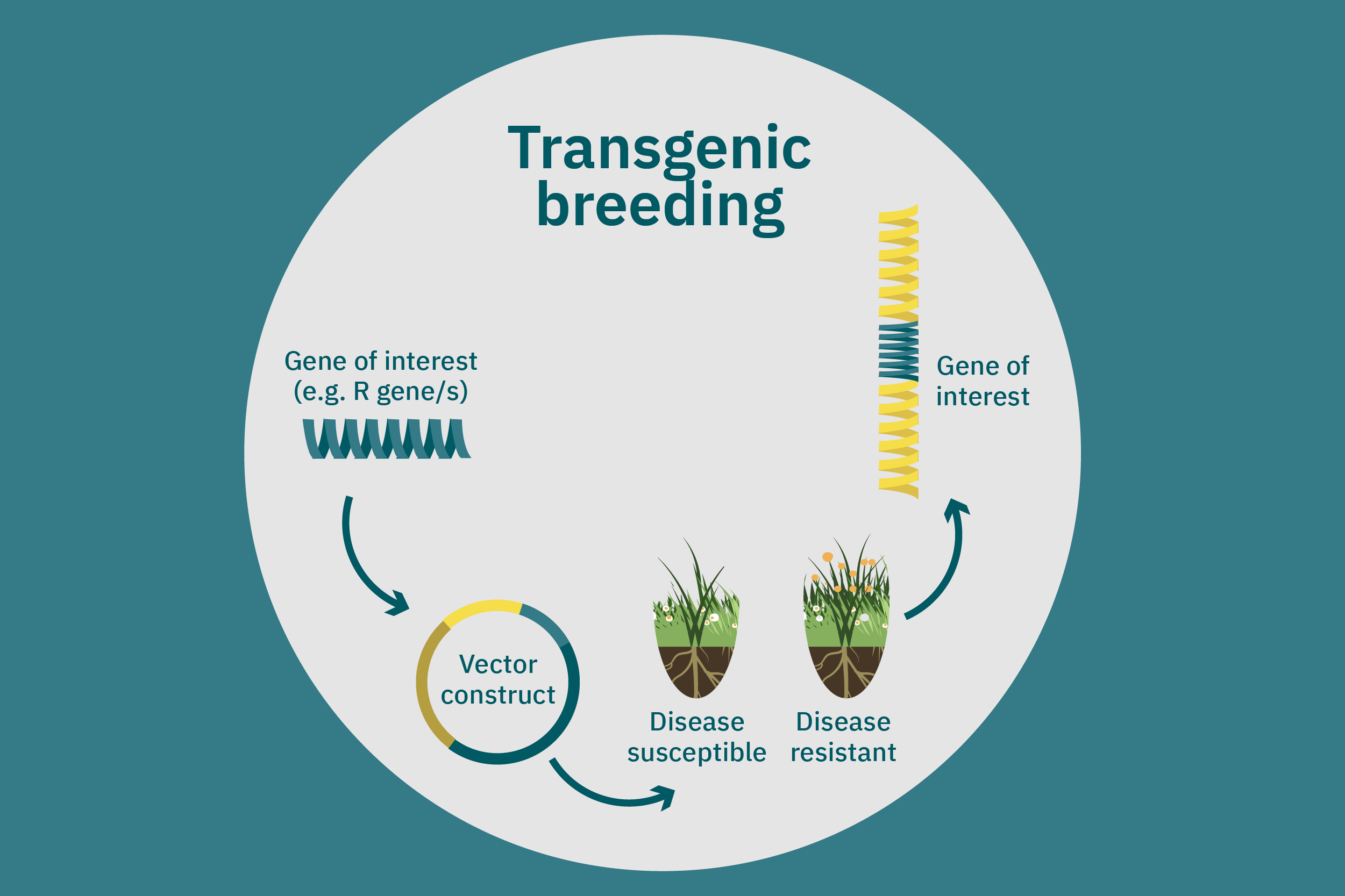
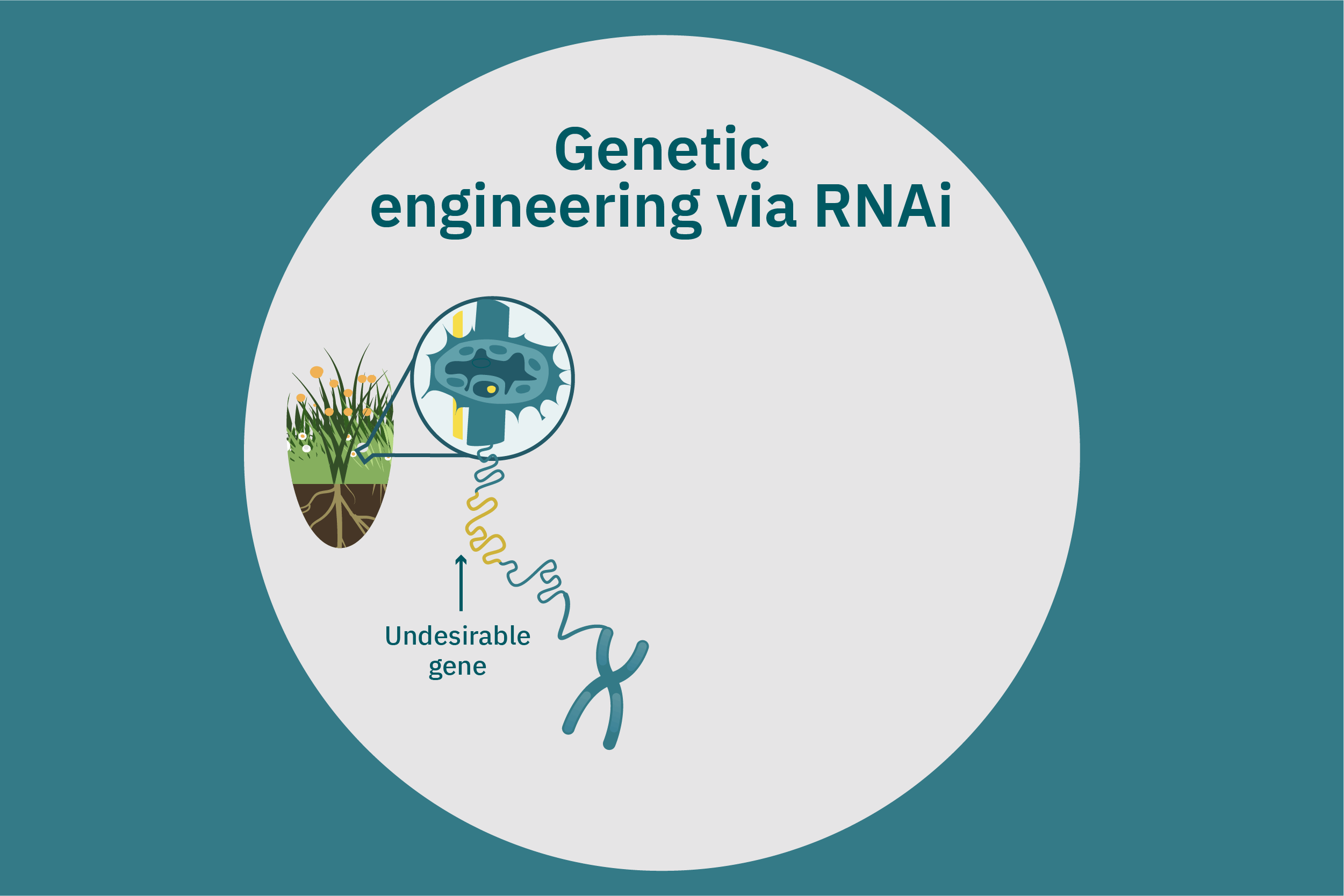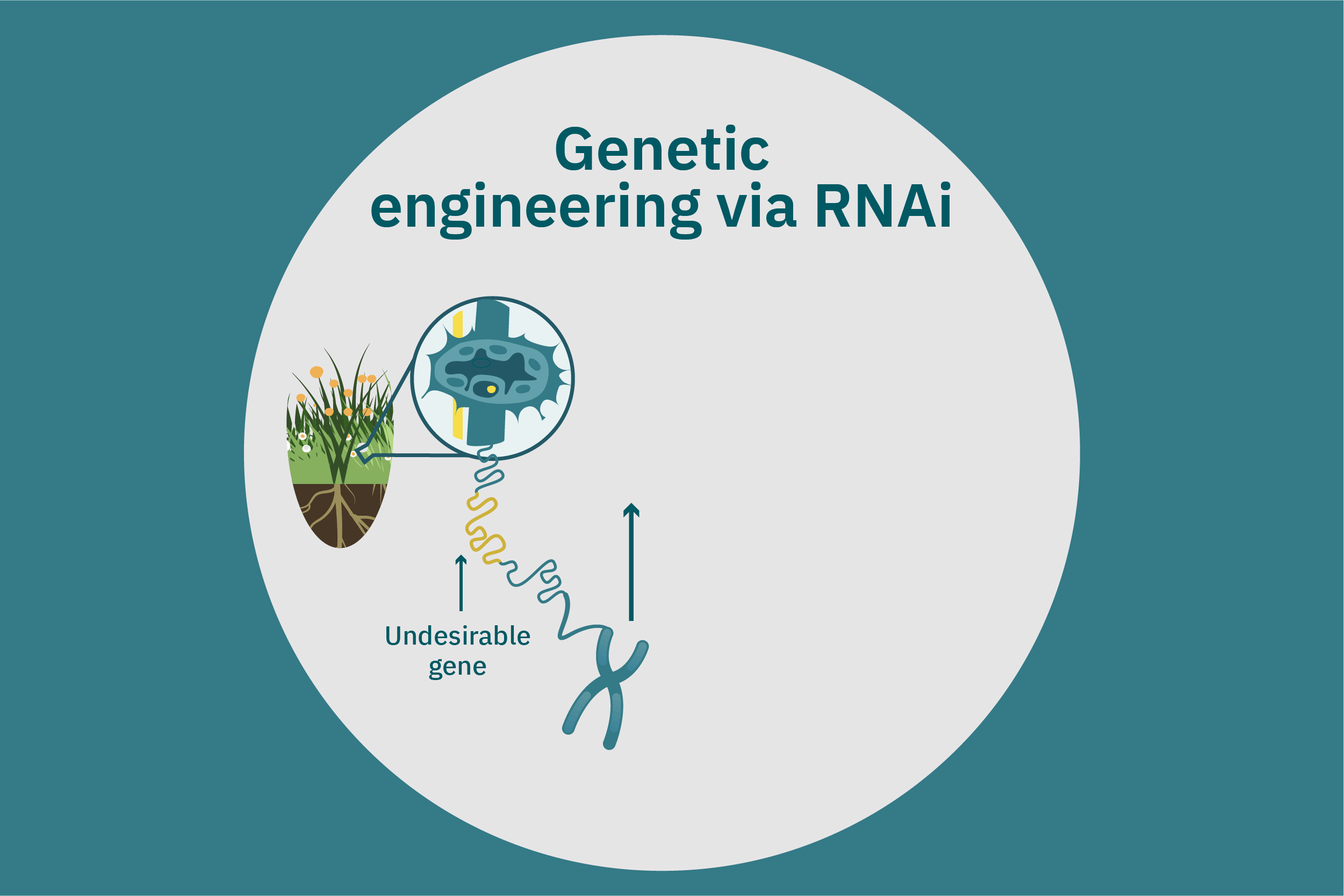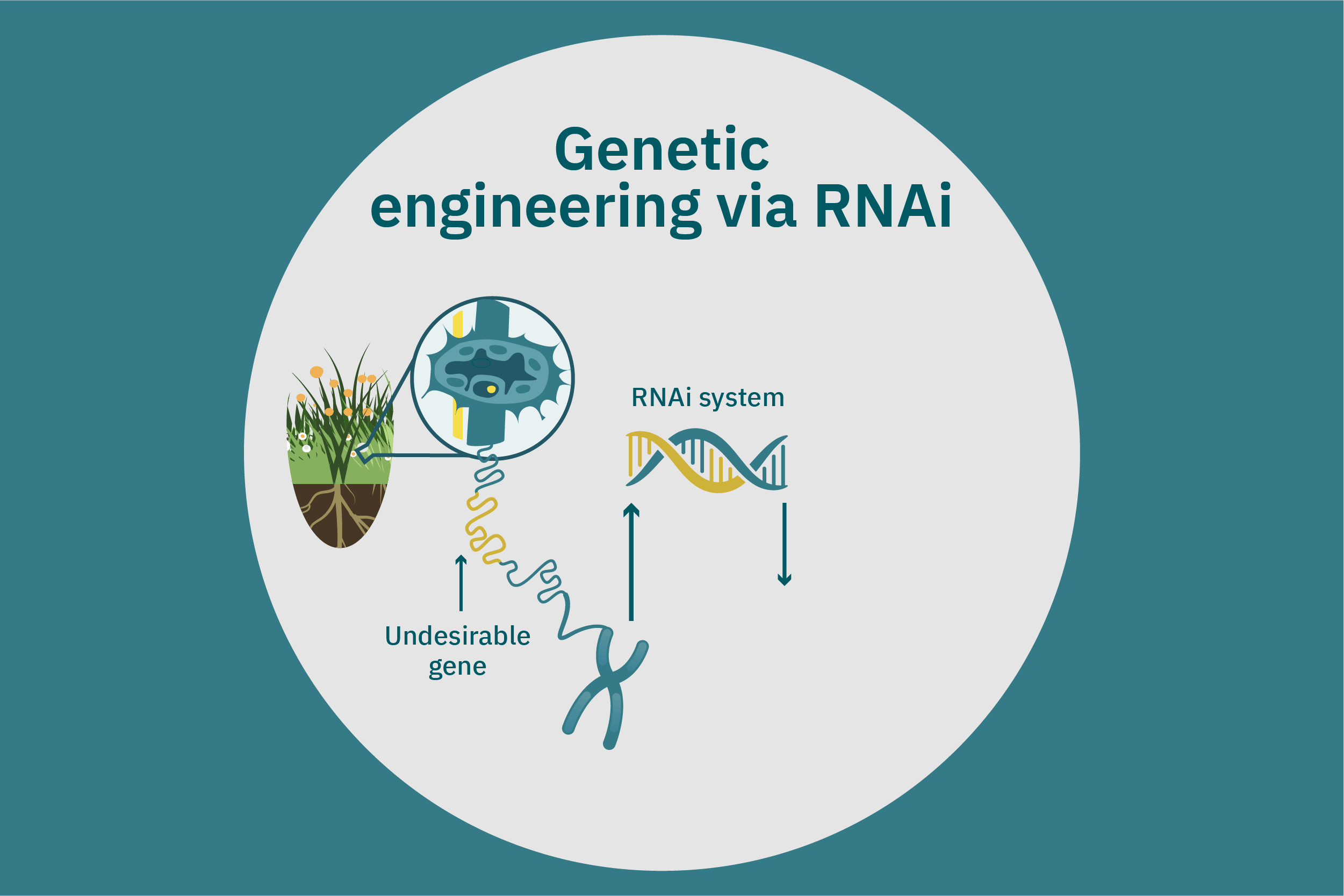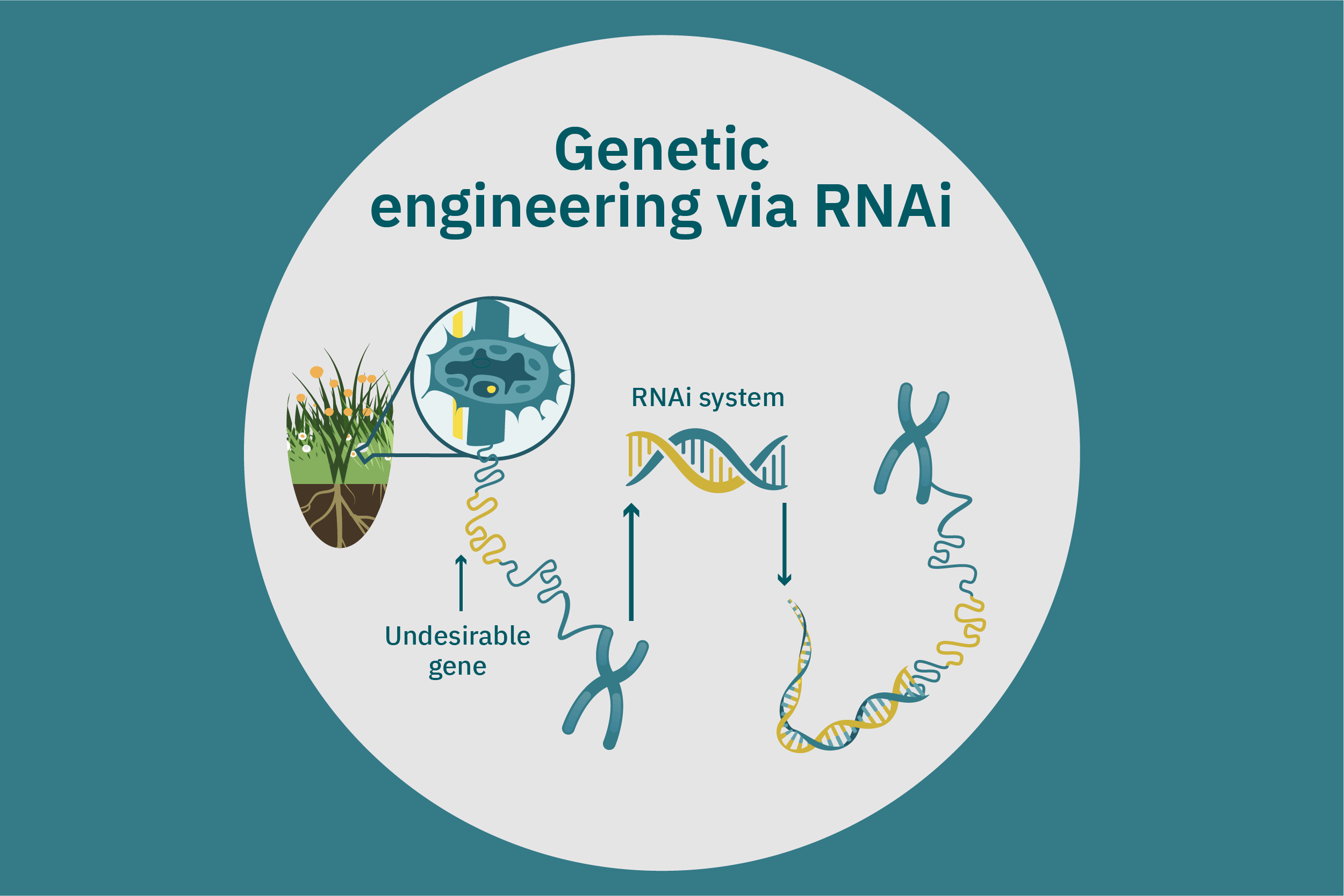
While conventional crop breeding can be viewed as the 50-50 transfer of genes from two crops into a new crop, genetic engineering transfers selected genes from one organism into a crop. Genes of interest are isolated from an organism and then incorporated into a vector to generate recombinant DNA. The vector is then inserted into plants using microinjection, a gene gun, or virus- or bacteria-mediated approaches. A common type of vector used for bacteria-mediated gene transformation is a plasmid with promoter and terminator regions. For example, to create potatoes with well-balanced essential AAs in potato proteins, Chakraborty et al. 2000 & 2010 used bacteria-mediated transformation. Expression plasmids with a gene that encoded for a well-balanced amaranth seed protein were constructed and mobilized into Agrobacterium tumefaciens bacteria. Potato stem segments were then incubated in a saturated culture of the plasmid-containing bacteria. The amaranth seed protein was successfully expressed in the cytoplasms and vacuoles of the potato plant cells, thus enhancing their AA composition. Other genetic techniques, such as RNA interference, can suppress undesirable genes (Abhary et al. 2015).
Precise genome editing innovations apply site-specific endonuclease enzymes to create a targeted, double-stranded cut in the host genome. Endogenous DNA repair machinery then repairs the sequence, sometimes including a transfer of new genes, depending on the repair method and application. There are various types of genetic editing tools, including zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and clustered, regularly interspaced short palindromic repeats-Cas9 (CRISPR-Cas9). ZFNs and TALENs have some limitations, as off-target effects can cause cytotoxicity. CRISPR-Cas9 is the newest of these technologies and has high specificity in crop breeding strategies (Ahmad et al. 2021). For example, CRISPR-Cas9 was used to reduce phytate concentration in corn (Liang et al. 2014), decrease the fatty acid content of camelina (Camelina sativa) seeds (Jiang et al. 2016; Morineau et al. 2016), and control starch branching by decreasing amylopectin and increasing amylose content in rice (Sun et al. 2017). Yield10 and Rothamsted are producing a CRISPR-edited camelina crop as a source for plant-based omega-3 fatty acids, a nutrient that has proved vital for alternative meat and seafood development. Rothamsted research was recently granted approval to conduct field trials of a CRISPR-edited wheat crop in the United Kingdom. CRISPR-edited crop trials are unprecedented in Europe, demonstrating the promise of the emerging technology’s use in food systems.
Genetic engineering can also be applied to “molecular farming,” or the use of plants as recombinant protein expression platforms for functional food ingredients. For example, as mentioned, Nobell Foods has applied genetically modified soybeans to produce casein protein. Additionally, Moolec Science uses soybean, pea, and safflower crops to produce bovine- and porcine-based proteins. See GFI’s solutions database for more information on leveraging crops as recombinant protein production hosts.
Note that optimizing protein fractions to enhance enriched protein powder functionality is an underexplored opportunity in crop breeding that can significantly impact plant-based meat products.
Comparing traditional breeding and genetic engineering
Choosing a crop breeding method depends on several factors: the trait of interest; the source, number, and location of genes that express this trait; properties of the crop; and considerations for downstream consumer preferences or regulatory hurdles associated with each technique. Traditional breeding is limited to naturally occurring variations and may introduce superfluous genes along with the targeted genes. On the other hand, genetic engineering can incorporate more versatile genes and be more precise, transferring only the desired gene to the desired location. It can also be faster, since a single generation is typically all that is required to introduce the desired trait. However, traditional breeding can be less time-consuming for more complex, multigene, or naturally occurring traits, especially in conjunction with speed breeding and other technologies. Genetic engineering is still gaining acceptance in some regions of the world, and development is often stalled due to consumer perceptions and government regulation (Anders et al. 2021; Turnbull et al. 2021). Registration for a new genetically engineered crop cultivar can take years in the United States. The European Union and some Latin American countries have even stricter regulations and lower consumer acceptance rates.
In some cases, producing functional components outside plants could be more efficient. Strain development could be performed much more rapidly in organisms such as fungi, microalgae, and other microbes than in crop plants due to their rapid doubling rate and relative ease of genetic manipulation. Precision fermentation is explored further on GFI’s science of fermentation web page. It is important to note that proteins formed through precision fermentation often lack posttranslational modifications, such as glycosylation, provided by the native hosts, so rigorously testing structure-function relations is especially important in this context. Host engineering in either case can alter the patterns of posttranslational modifications to more accurately reflect the desired configuration, but this process adds time and complexity.
Bioinformatics for alternative proteins
There is a need to connect protein (and other raw ingredient) functionality in plant-based meat to the sequence and structure of the ingredient and the associated phenotypes and QTLs of the protein’s crop source. GFI’s solutions database includes more information on protein sequence, structure, and functionality databases. This concept could be expanded to include artificial intelligence, machine learning, and bioinformatics to understand the genetic contributions to protein sequence and structure. Bahmani et al. 2021 recently applied proteomics to investigate barley proteins and their biological roles. The authors expressed that additional research is needed to guide breeding for more functional food proteins. More open-access databases and tools to interpret biological data would result in a more sophisticated understanding of proteins, thus producing more plant-based meat innovations.